-
OPEN positions !
We are looking for a person with experience in bioinformatics, computational biology or data analytics.
If interested, please email to : linas.mazutis at bti.vu.lt
Our current research topics include:
|
 |
1. Single-cell transcriptomics, genomics and epigenomics
Simultaneous profiling of the genome, epigenome and transcriptome of the same cell has been a dream of many biologists for decades. Technological advances over the last few years have brought us closer to this reality, by enabling single-cell transcriptomics, epigenomics, methylomics, genomics. However, the integration of these individual modalities into a unified single-cell multi-omics profiling has proven a significant challenge. Although, latest developments have achieved dual-omics (joined-profiling of transcriptome and epigenome), yet the ultimate goal in single-biology field is to integrate all layers of genetic and epigenetic information of individual cells. Simultaneous -omics studies on millions of single-cells are especially challenging not only due to technical difficulties associated with clinical sample processing and efficient single-cell isolation, but also due to complex, multi-step and sequential biochemical reactions that need to be conducted on miniscule amounts of biomaterial. To address this challenge we are developing new molecular barcoding tools to simulteniously profile multi-ome (genome, epigenome and transcriptome) of the same cell in a high-throughput and affordable cost.
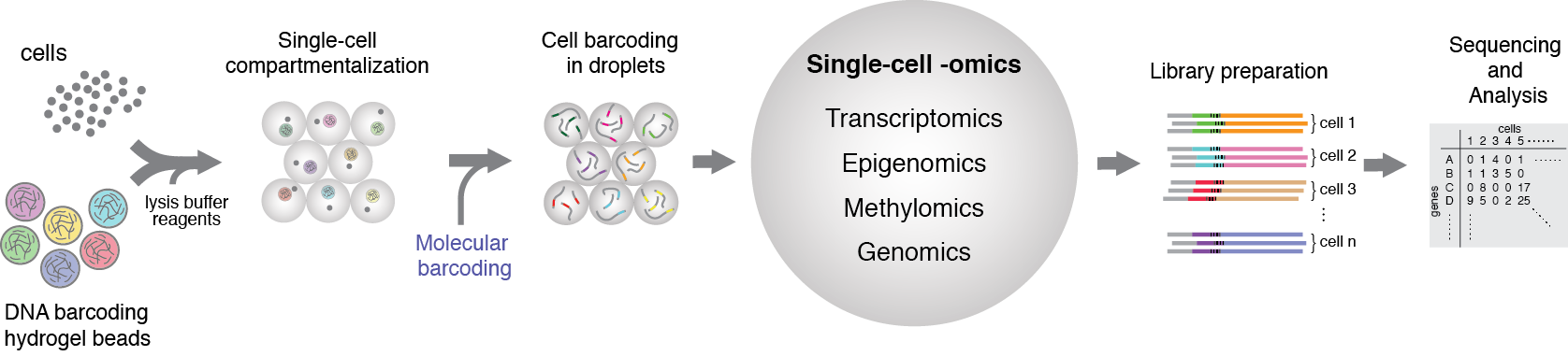
Figure 1. General schematics of the single-cell multiome approach that we are currently developing in our group. The idea is build on isolation of individual cells in microfluidic droplets together with suitable reagents for molecular barcoding of transcriptome, epigenome, genome and methylome. Leveraging unique features of microfluidics technology and applying suitable biochemical reagents it is possible to achieve indexing of individual cellular modalities simultaneously.
2. Single-cell barcoding and sequencing using droplet microfluidics
Deciphering the unbiased composition of heterogeneous cell populations requires innovative techniques that could capture not hundreds but thousands of single-cells and to do so in a high-throughput manner and affordable cost. This work is built on idea of isolating individual cells into microfluidic droplets carrying RNA/DNA barcodes, and assay reagents. Because individual cells are compartmentalized in drops, cell genetic make up and contents can be accessed. For single-cell studies our approach provides unprecedented scalability, significantly increased throughput, minimal sample loss, reagent savings and large experimental flexibility. Using this technology we are interested in understanding the genetic and epigenetic factors that are responsible for cellular heterogeneity and inheritance.
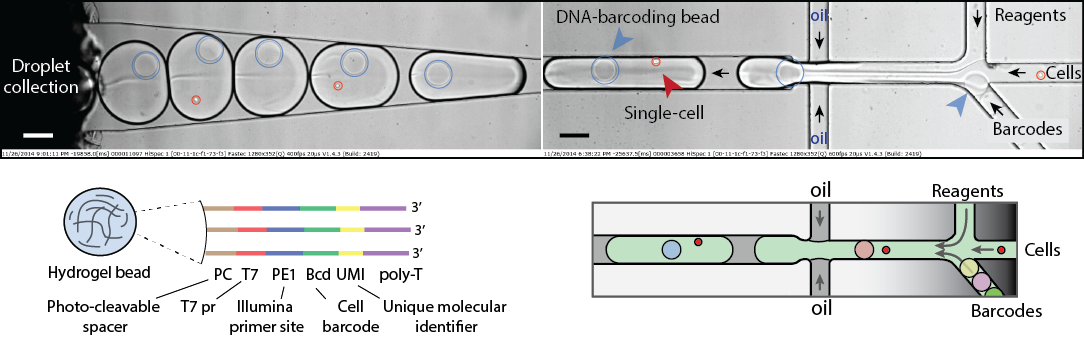
Figure 2. Single-cell transcriptomics. Heterogenious mix of cells is encapsulated into droplets together with reverse-transcription enzyme, lysis mix, and hydrogel beads carrying barcoding primers. After encapsulation cells are lysed, primers are released from the beads and cDNA is tagged with a barcode during reverse transcription reaction. Once cDNA synthesis is complete, droplets are broken and barcoded material from all cells is amplified for next-gen sequencing. Read to learn more
3. High-throughput screening of antibody secreting cells
In this project, we use droplet microfluidics platform for high-throughput single-cell screening of cells that produce therapeutic antibodies or biomolecules of biomedical interest. Cell compartmentalization into microfluidic droplets together with capture beads and barcoded DNA primers enables us a direct establishment of the linkage between the genotype (genes or mRNA) and phenotype (binding, regulatory or activity of secreted proteins). Due to increased local concentration inside droplets the amount of secreted antibodies will significantly reduce the time window necessary for the assay. We aim at developing a discovery pipeline for the quantitative high-throughput antibody phenotyping without loosing the original heavy-light chain pairing, which would be a significant advantage over other technologies. Our technological approach provides a unique way to identify the primary sequence of heavy and light IgG genes encoding functional monoclonal antibodies directly from single-cells, without a need to perform gene cloning, hybridoma construction or cell immortalization.
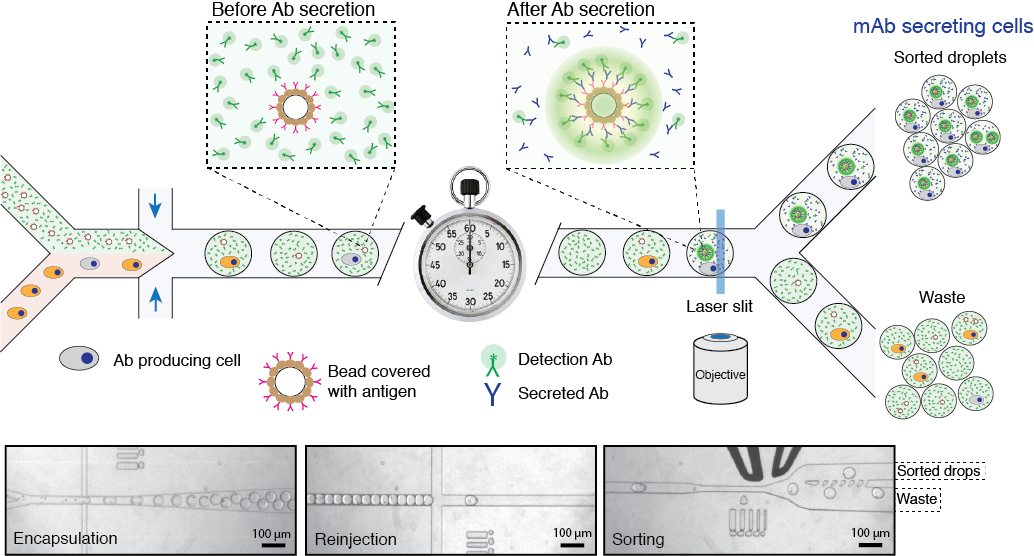
Figure 3. Schematics of microfluidics platform for screening and sequencing single-cells. A mixture of cells is loaded into droplets together with assay reagents and beads carrying capture probe (antigen). The cells secreting antigen specific Ab will be detected by fluorescent sandwich assay on bead surface and sorted. The sorted cells will be lysed and mRNA encoding heavy and light chains are captured and sequenced. Alternatively, the sorted cells can be recovered directly into a growth media for further cultivation, propagation and analysis. Read to learn more
4. in vitro directed evolution using artificial cells
Darwinian evolution is a powerful algorithm that has given rise to the functionally diverse set of proteins present in all living systems. Repetitive rounds of mutation, selection and amplification have optimized nature’s catalysts, the enzymes, to an extraordinary degree, for example. Biocatalysts are typically much more efficient than their man-made counterparts, often working close to the diffusion limit. A better understanding of how new enzymes evolve consequently remains an important and challenging task for both academic and industrial needs. Although the active sites of natural enzymes are highly complex, making the design of new or promiscuous protein functions difficult, computation has recently emerged as a potentially powerful strategy for creating protein catalysts with tailored activities and specificities. Because the activities of such artificial enzymes are still modest, we plan to explore the feasibility of optimizing them by combining the latest advances in droplet-based microfluidics technology with the methods of directed evolution. Specifically, two complimentary groups, one with expertise in microfluidic systems and another with expertise in directed protein evolution, will join forcess to investigate mechanisms and strategies for optimizing several computationally designed esterases. By applying droplet-based microfluidics technology, in which biochemical reactions are performed inside micrometer size vessels, we will have a powerful tool enabling large number of libraries (>10^6) to be screened at ultra-high-throughput rates and using conditions that are incompatible with in vivo systems.

Figure 4. Concept of directed evolution approach in droplet microfluidics. Droplets containing single-genes with all ingredients necessary for in vitro expression will serve as artificial cells that can be selected for a desirable phenotype under conditions that are not feasible in living systems. Library of mutated genes is encapsulated such that, statistically, the majority of droplets contain no more than one gene per droplet. After IVTT reaction the droplets are fused with a second type of droplet containing a fluorogenic substrate and other molecules, salts, and reagents needed to impose the selection pressure. After mixing and incubation, the fluorescence of each droplet is recorded. Droplets with fluorescence above a threshold are sorted by means of dielectrophoresis and the genes contained therein are amplified using PCR. The selected genes can be either characterized, re-selected, or mutated for subsequent round of directed evolution.
5. Droplet microfluidic tools for biological and biomedical applications
Our group has long history of experience at developing novel microfluidic tools for precise manipulation and analysis of biological reactions at pico and nanoliter range volumes. We design, model, fabricate, characterize and implement microfluidic systems and devices for myriad of biological and biomedical applications for our colleagues and industrial partners. We build experimental platforms based on integration of microfluidics, mechanical, electrical and optical modules eventually enabling precise control over experimental conditions hardly feasible in bulk. Using droplet microfluidic devices we provide significantly reduced assay volumes for virtually any biological assay and as a result big cost savings for biochemical reagents and compounds. We are developing microfluidic devices and chips for digital DNA/RNA analysis, single-cell and biomedicine applications.
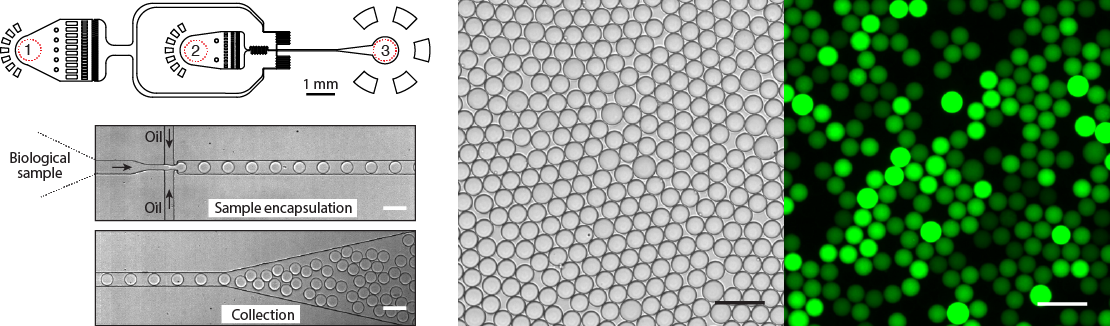
Figure 5. Design and operation of the droplet microfluidics device. Almost any biological sample (DNA, cells, proteins) can be encapsulated into pico- or nano-liter volume droplets at >10.000 droplets per second. Typical microfluidics device has (1) the inlet for the continuous phase, (2) the inlet for biological sample, and (3) the droplet collection outlet. Bright field and fluorescence images of an emulsion after DNA amplification reaction. Droplets containing amplified DNA exhibit green fluorescence, whereas droplets lacking a template are dark. Scale bars, 50 um.
6. Bacterial genomics and transcriptomics
Single bacterial genomics holds a great promise of improving our understanding of the microbiology world, discovery of new enzymatic activities, or identifying the antibiotic resistance amongst others. However, performing single genome amplification (SGA) of individual bacterial in water-in-oil emulsions can be hindered by the preceding cell lysis step, leading to inefficient or non-uniform DNA amplification or, in case of hydrogel beads, significant cell loss during gel solidification. Bacterial lysis steps can be particularly problematic for single-step isothermal nucleic acid amplification methods (e.g., multiple displacement amplification [MDA]) or working with gram-positive bacteria, which are known to be much more resistant to thermolysis. To circumvent these limitations we have recently introduced a method for producing biocompatible capsules containing a thin, semi-permeable shell. The shell acts as a passive sieve — retaining large molecular weight compounds such as genomic DNA, while allowing smaller molecules (such as proteins) to diffuse through. We use an aqueous two-phase system (ATPS) composed of dextran and acrylate-modified polyethylene glycol to generate the biocompatible hydrogel particles apply it for whole genome amplification of gram-negative and gram-positive bacterial cells. Capsules readily sustain multiple pipetting steps when performing complex biochemical reactions, and, generate higher genome amplification yields.
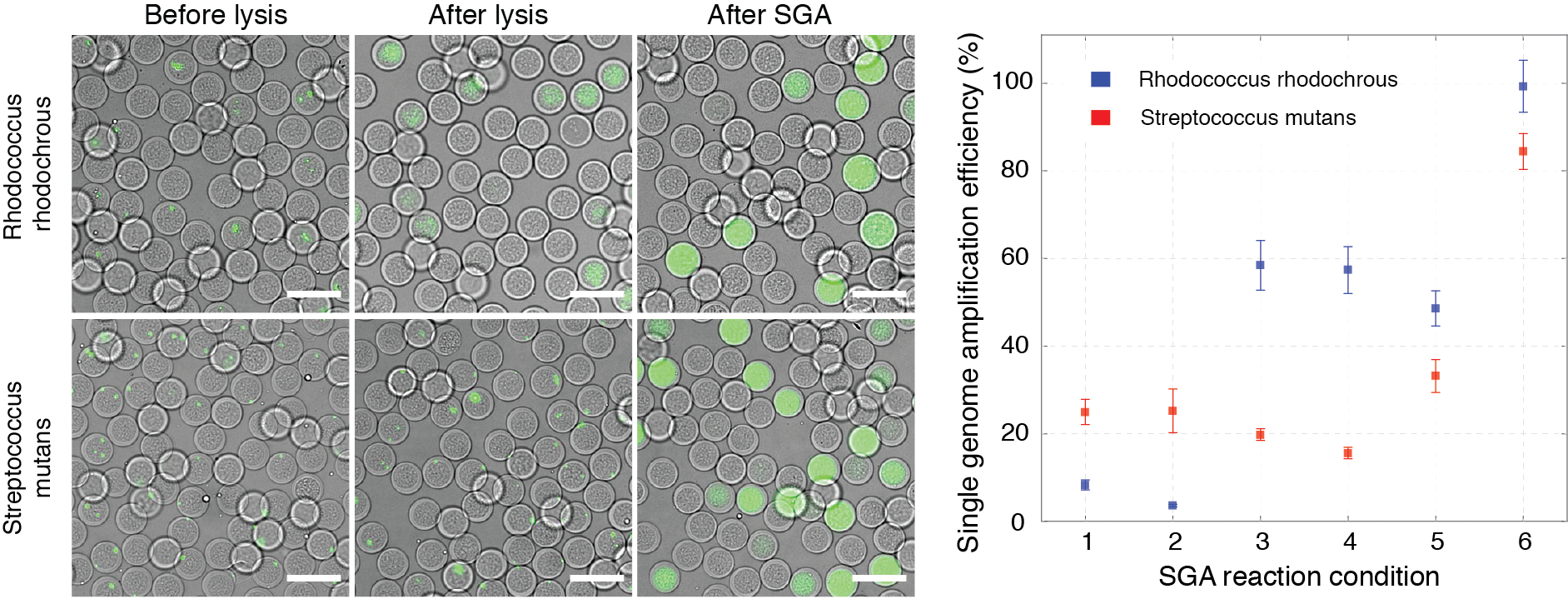
Figure 6. Single genome amplification on individual microbial cells is more efficient in semi-permeable capsules than either hydrogel beads or droplets. Proteinase K and SDS treatment yielded the largest SGA on individual gram-positive cells using semi-permeable capsules. On the left, digital images showing capsules stained with DNA binding dye (SYBR Green I) before lysis, after lysis, and after SGA. On the right, single genome amplification efficiency is expressed as a percentage of observed over expected DNA amplification reactions of gram-positive bacteria lysed and processed at different conditions. Scale bars, 50 μm. Read to learn more
7. DNA supramoleular crystals and their use
Compartmentalization and amplification of single DNA molecules inside nano- or pico-liter sized wells and droplets opens new opportunities for biomedical and biological sciences. The most common method of amplifying DNA in a sample involves the polymerase chain reaction (PCR). Yet, amplification of long (>1 kb) single-molecule templates is often inefficient, leading to reduced reaction yields and biases. DNA amplification under isothermal reaction conditions has been shown to generate large amounts of material from a single-copy DNA template. Moreover, the ability to amplify DNA and then express proteins from the clonally amplified template would greatly increase the scope of potential applications. We employ droplet microfluidics devices to encapsulate and convert single DNA molecules into condensed DNA nanoparticles by a multiple displacement amplification (MDA) reaction driven by phi29 DNA polymerase. The condensed DNA nanoparticles comprise up to ~10.000-100.000 copies of clonally amplified DNA template. Intriguingly, we found that inorganic pyrophosphate, produced during isothermal DNA synthesis, and magnesium ions are a prerequisite for DNA condensation into the crystalline-like globular structures. This process is enhanced when the DNA amplification reaction is performed inside droplets, which can be attributed to the confined volumes and spatial accumulation of the reaction products. We have demonstrated the biological functionality of the DNA nanoparticles, by applying them in in vitro transcription-translation reactions and observed improved protein expression yields relative to standard assay conditions.
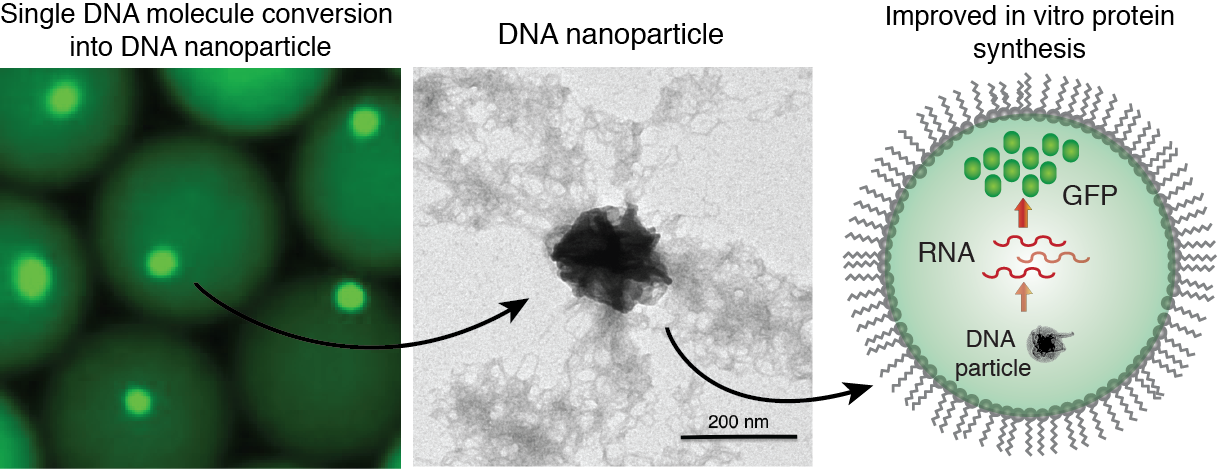
Figure 7. Generation of DNA-Mg-PP particles. DNA nanoparticle formation induced by inorganic pyrophosphate (PP) and magnesium ions during a phi29-catalyzed DNA polymerization reaction. TEM image of the DNA-Mg-PP particle. The use of DNA for efficient gene expression and protein synthesis in vitro. Read to learn more
