|
SONATA JARMALAITĖ |
|
RESEARCH OVERVIEW
The incidence of cancer is continuing to rise. The current COVID-19 pandemic reduced cancer screening activities, restricted timely access to clinical service and negatively impacted early cancer detection. Thus, in 2021 the European Union is planning to start an implementation of new Cancer Plan with the main goal – to minimise cancer mortality.
During the last decade, the increased understanding of genetic alterations in tumours has personalised cancer treatment through the usage of molecular diagnostic tools. Molecular tests have been developed in order to facilitate both the diagnosis of the disease and the selection of the most effective treatment scheme, as well as to avoid unnecessary interventional procedures for the patient. Genomic data are used for the identification of new drug targets of personalized cancer therapy. Future progress in oncology is feasible through further development and implementation of personalised medicine means.
Using a variety of genome-wide and target-oriented methodologies [1], our group aims at the (epi)genetic characterization of various human tumours and the development of molecular biomarker systems for cancer detection, molecular classification, prognosis and early identification of resistance development [2–4]. Focusing on altered DNA methylation, we have recently proposed biomarker panels for prostate cancer detection and more accurate characterization by using liquid biopsy samples [3]. Genetic and epigenetic analyses were used to identify novel players of renal carcinogenesis. Some of metallothionein genes were shown as specifically hypermethylated in renal cell carcinomas and showed significant associations with clinical variables of the disease [2]. In collaboration with the Institute of Biochemistry and National Cancer Institute of Lithuania, we continued the investigation of molecular mechanisms of combined action of dichloroacetate and salinomycin for the development of novel solutions in cancer therapy [4].
SELECTED PUBLICATIONS
- Daniunaite, K., Jarmalaite, S., Kriukiene, E. Epigenomic technologies for deciphering circulating tumor DNA. Curr Opin Biotechnol. 2018, 55: 23–9. http://doi.org/10.1016/j.copbio.2018.07.002.
- Maleckaite, R., Zalimas, A., Bakavicius, A., Jankevicius, F., Jarmalaite, S., Daniunaite, K. DNA methylation of metallothionein genes is associated with the clinical features of renal cell carcinoma. Oncol Rep. 2019, 41: 3535–44. https://www.spandidos-publications.com/or/41/6/3535.
- Bakavicius, A., Daniunaite, K., Zukauskaite, K., Barisiene, M., Jarmalaite, S., Jankevicius, F. Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin Epigenetics. 2019, 11: 115. https://clinicalepigeneticsjournal.biomedcentral.com/articles/10.1186/s13148-019-0716-z.
- Skeberdytė, A., Sarapinienė I., Krasko J. A., Barakauskienė, A., Žilionytė, K., Prokarenkaitė, R., Sužiedėlis, K., Bukelskienė, V., Jarmalaitė, S. Salinomycin and dichloroacetate synergistically inhibit Lewis lung carcinoma cell proliferation, tumor growth and metastasis. Biochem Biophys Res Commun. 2020, pii: S0006-291X(20)30058-9. https://www.sciencedirect.com/science/article/pii/S0006291X20300589.
RESEARCH ACTIVITIES in 2020
Predictive Value of Germline DNA Damage Repair Gene Mutations in Metastatic Castration-Resistant Prostate Cancer
Prostate cancer (PCa) is the second most prevalent oncologic disease and the fifth leading cause of cancer-related deaths among men worldwide. PCa is a heterogeneous and dynamic disease, which usually progresses to metastatic castration resistant PCa (mCRPC) – the highly lethal form of PCa. Recent data suggest that mCRPC may carry the germline or sporadic mutations in DNA damage response (DDR) pathway genes, and this cancer is commonly identified in families with a high predisposition to the breast and ovarian cancer. To determine the relationship between DDR status and clinical response to therapy as well as survival outcomes, 149 leukocyte samples from patients with mCRPC were evaluated. The number of known mCRPC predisposition genes is limited and includes BRCA1, BRCA2, CHEK2, ATM, NBN as well as some other genes. Our results demonstrated that pathogenic germline mutations were detected in 15.4% of the patients (Fig. 1). Next generation sequencing-based studies can uncover the complex genomic landscape of mCRPC and identify novel targets for efficient anticancer therapies.

Fig. 1. Combined gene mutation analysis data from 23 patients. Gleason score, metastasis place, tumour stage and gene mutation type is shown.
Androgen Receptor Variant Analysis for Predicting Response to Abiraterone Acetate in Patients with Castration Resistant Prostate Cancer
Androgen receptor (AR-FL) and AR variants (AR-V1, -V3, and -V7) were evaluated in 185 serial blood samples, prospectively collected from 102 patients with castration resistant prostate cancer (CRPC) before and during abiraterone acetate (AA) therapy via reverse transcription quantitative PCR. AR-FL was present in all the samples, while AR-V1, AR-V3, AR-V7 were less frequent, and at least 1 of them was detected in 17%, 55%, 65% and 81% of CRPC patients’ blood samples, respectively. The highest amount of AR-V1 was found in the blood of patients whose response time was short and medium in comparison to extended (Fig. 2). Patients with a higher level of AR-FL and/or AR-V1 had the shortest progression-free survival and overall survival (p<0.0001). Blood circulating AR-FL or AR-V1 can serve as blood based biomarkers for identification of the primary resistance to AA and the tool to monitor de novo resistance development during AA treatment.
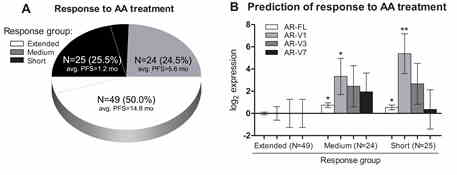
Fig. 2. Prediction of response to AA treatment by level of AR transcripts at baseline and variation of AR transcript level in CRPC blood during treatment. A – PFS of patients treated with AA grouped by response time. B – Prediction of response to AA therapy from AR-FL and AR-Vs, detected in blood samples collected before treatment initiation. * – p<0.050, ** – p<0.001
Frequent DNA Methylation Changes in Cancerous and Noncancerous Lung Tissues of Smokers with Non-Small Cell Lung Cancer
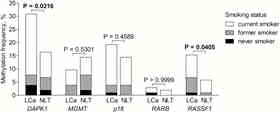
Lung cancer (LCa) is the leading cause of cancer-related deaths. Smoking is one of the major LCa risk factors, and tobacco-related carcinogens are potent mutagens and epi-mutagens. The present study evaluated DNA methylation status of tumour suppressor genes (TSG) DAPK1, MGMT, p16, RASSF1 and RARB in paired LCa and noncancerous lung tissues (NLT) of 104 patients, 90 of whom were smokers or ex-smokers. Methylation of at least one gene was detected in 59% of LCa samples and in 39% of NLT. The levels of DNA methylation were higher in LCa than NLT in most of the analysed CpG positions (Fig. 3). More frequent methylation of at least one gene was observed in LCa samples of ever smokers (63%) as compared with never smokers (36%). In the ever smokers group, methylation of the genes also occurred in NLT, but was rare or absent in the samples of never smokers. Therefore, we conclude that DNA methylation changes are widespread in cancerous and NLT of smokers creating an epigenetically altered field for cancer development and further spread.
B77–7
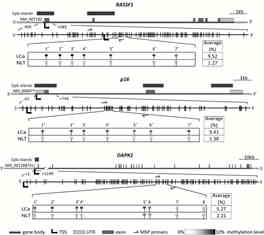
Fig. 3. DNA methylation analysis of TSG’s in lung tissues.
A - Methylation frequencies of the genes in LCa and NLT. Colours indicate patients’ smoking status. B – Quantitative methylation analysis of particular CpG positions at the regulatory regions of the genes.


