|
SAULIUS SERVA |
|
RESEARCH OVERVIEW
The Totiviridae family dsRNA viruses from the Saccharomycetaceae family yeast are ubiquitous yet poorly understood benign inhabitants of the host. In our lab, they are being investigated by means of molecular biology techniques. The impact of the dsRNA viruses uncovered by genomic, transcriptomic, proteomic and phenomic analysis is interpreted as a model framework for establishing the universal mechanisms behind any virus of interest, in such a way creating a paradigm network for virus-host interactions. We aim at the understanding of intra- and extracellular relations of yeast dsRNA viruses in order to elucidate the evolutionary pathways of these viruses and reveal the ultimate principles of distribution within an ecosystem [1].
Nucleoside and nucleotide-based antivirals constitute the essence of modern high-efficacy antiretroviral HIV treatment. Once a revolutionary approach upon discovery, nowadays it suffers from an emerging resistance and multiple side effects due to life-long administration. Recently, innovative and more advanced measures against genuine retroviral replication enzymes have been proposed and substantiated. The aim of our research is to develop compounds active at the level of a catalytic cycle of retroviral replication enzymes, linking an exclusive specificity and efficacy into a binding approach.
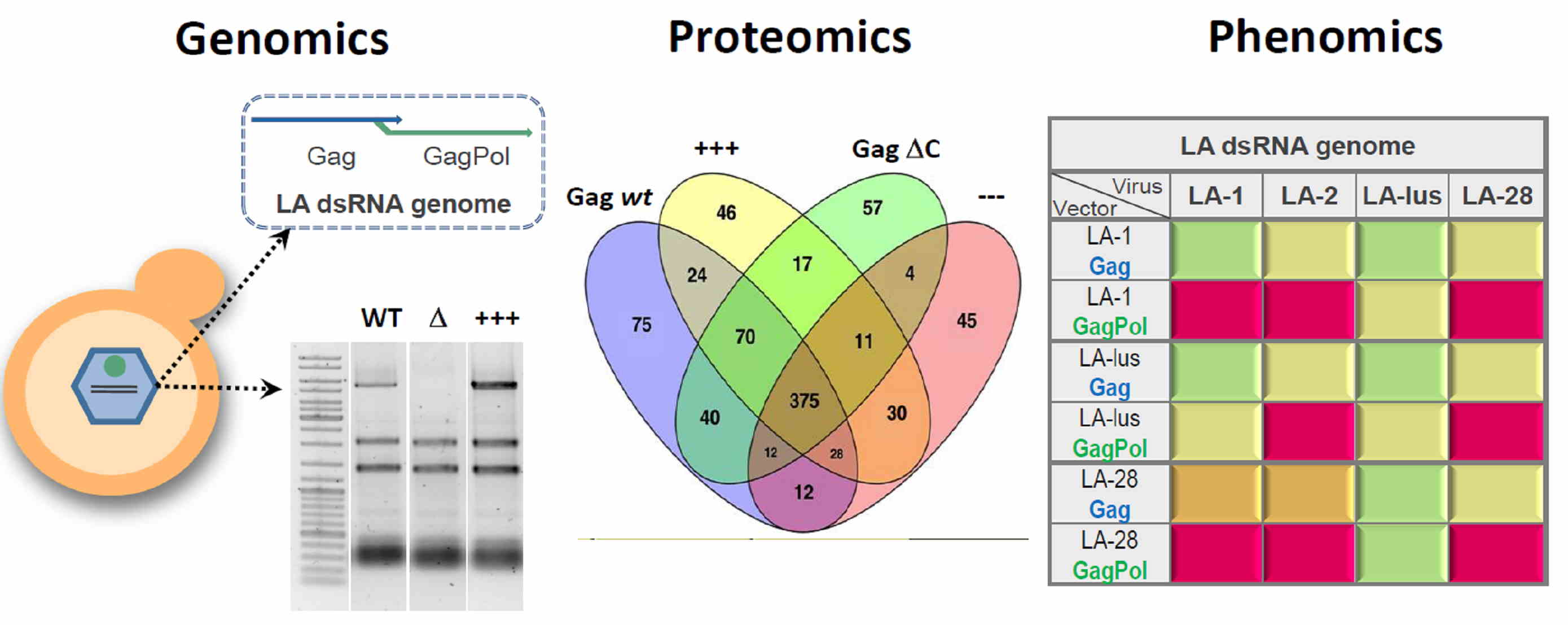
Our team focuses on systems biology approaches to address the interactions of yeast double-stranded RNA Totiviridae viruses with the host cell. Basing on a virus genome cloning technique, developed in our lab [2], constituent genes of a virus genome were re-introduced into model hosts to manipulate the phenotype conferred by the virus. We were able to either achieve a complete clearing of the target virus or boost the synthesis of the viral genome, making it the most prevalent form of an individual RNA molecule in a cell. The developed techniques allowed us to perform a transcriptomic and proteomic analysis, aimed at understanding the molecular mechanisms behind the establishment of Totiviridae viruses in host cell [3].
To create novel universal antiviral compounds, we took advantage of the catalytic mechanisms of viral polymerases. In particular, the catalytic flexibility of reverse transcriptases from HIV and M.MuLV were exploited to prepare and investigate the conjugates of nucleotide and small molecule inhibitors. We demonstrated the feasibility of altering the action of a polymerase, forcing a shift from the processive to the distributive mode [4]. The conformational alterations of productive complexes were postulated to determine the impaired turnover of the target enzymes, in such a way ensuring selectivity among a variety of cellular polymerases.
SELECTED PUBLICATIONS
- Vepštaitė-Monstavičė, I., Lukša, J., Konovalovas, A. et al. Saccharomyces paradoxus K66 killer system evidences expanded assortment of helper and satellite viruses. Viruses. 2018 Oct 16, 10(10).
- Grybchuk, D., Akopyants, N. S., Kostygov, A. Y. et al. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc Natl Acad Sci U S A. 2018 Jan 16, 115(3): E506–E515.
- Ravoitytė, B., Lukša, J., Yurchenko, V., Serva, S., Servienė, E. Saccharomyces paradoxus transcriptional alterations in cells of distinct phenotype and viral dsRNA content. Microorganisms. 2020 Nov 30, 8(12): E1902.
- Mikalkėnas, A., Ravoitytė, B., Tauraitė, D. et al. Conjugation of phosphonoacetic acid to nucleobase promotes a mechanism-based inhibition. Journal of Enzyme Inhibition and Medicinal Chemistry. 2018, 33(1): 384–389.