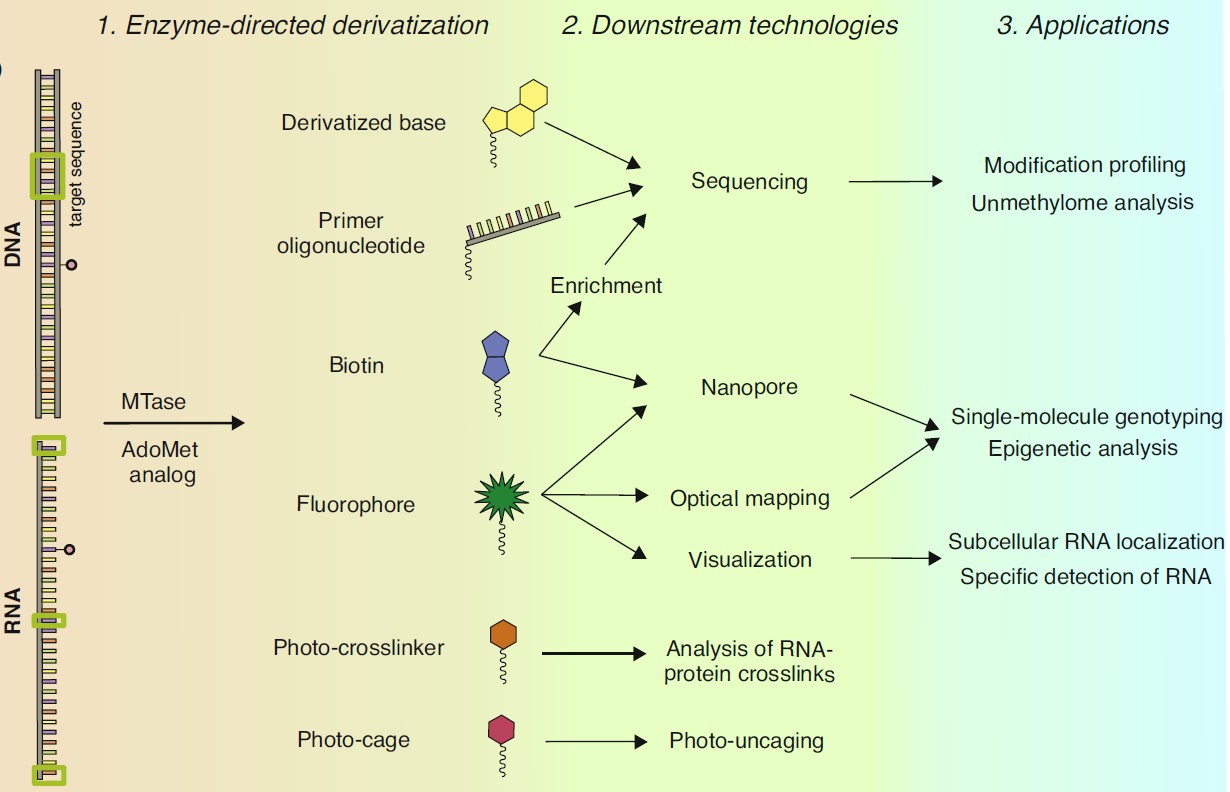
Molecular Tools for Targeted Covalent Derivatization of DNA and RNA
Nucleic acids are linear biopolymers comprised of four major types of building blocks encoding the genetic blueprint of life. Analysis of such largely uniform biomolecules can be facilitated by targeted installation of suitable bioorthogonal reporter tags. Among the variety of enzymes involved in nucleic acids metabolism, AdoMet-dependent methyltransferases (MTases) uniquely combine two features required for targeted labelling: recognition of a specific target and its covalent modification. To unlock the technological potential of these enzymes we seek to repurpose them for the transfer of pre-derivatized (extended) versions of the methyl group. A series of synthetic analogs of the AdoMet cofactor were developed that allowed MTases to tag DNA and RNA with extended moieties [1], inspiring the appearance of the mTAG technology (methyltransferase-directed Transfer of Activated Groups) for targeted covalent derivatization and labelling of DNA and RNA [2,3]. Beside their ability to transfer covalent labels on cytosine base in DNA, MTases have been devised to perform atypical C-C bond cleavage reactions, leading to removal of the oxidized one-carbon moieties (hydroxymethyl and carboxyl) and formation of unmodified cytosine in DNA [4]. By combining these MTase-promoted reactions, we develop novel tools for the analysis of epigenetic DNA modifications genome-wide, which could open new avenues in genomic research, diagnostics, and bionanotechnology.
Advanced Methods for Epigenome Analysis
Epigenetic regulation in vertebrates involves chemical variation of one-carbon groups of cytosine residues in CpG dinucleotides by enzymatic production of 5-methylcytosine followed by its oxidized forms 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxylcytosine. Genomic distribution of these modified cytosines varies in different cell types, environmental conditions and disease states and is associated with many biological processes such as embryogenesis, establishment of cell identity, and development of pathological conditions, including cancer. However, research into the epigenetic regulation is hampered by limitations of available analytical techniques. Recently, we proposed a high-resolution economical technique named TOP-seq, which exploits non-homologous priming of the DNA polymerase at covalently tagged CpG sites [5]. Using mTAG and other covalent DNA labelling technologies, we develop new experimental approaches for profiling cytosine modifications genome-wide for epigenome studies and improved diagnostics.
SELECTED PUBLICATIONS
- Tomkuvienė, M., Kriukienė, E., Klimašauskas, S. DNA labeling using DNA methyltransferases. Adv. Exp. Med. Biol. 2016, 945: 511–535.
- Kriukienė, E., Labrie, V., Khare, T., Urbanavičiūtė, G., Lapinaitė, A., Koncevičius, K., Li, D., Wang, T., Pai, S., Gordevičius, J., Wang, S. C., Petronis, A., Klimašauskas, S. DNA unmethylome profiling by covalent capture of CpG sites. Nat. Commun. 2013, 4: 2190.
- Osipenko, A., Plotnikova, A., Nainytė, M., Masevičius, V., Klimašauskas, S., Vilkaitis, G. Oligonucleotide-addressed covalent 3’-terminal derivatization of small RNA strands for enrichment and visualization. Angew. Chem. Int. Ed. 2017, 56: 6507–6510.
- Liutkevičiūtė, Z., Kriukienė, E., Ličytė, J, Rudytė, M, Urbanavičiūtė, G, Klimašauskas, S. Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases. J. Am. Chem. Soc. 2014, 136: 5884–5887.
- Staševskij, Z., Gibas, P., Gordevicius, J., Kriukiene, E., Klimašauskas, S. Tethered oligonucleotide-primed sequencing, TOP-seq: a high-resolution economical approach for DNA epigenome profiling. Mol. Cell. 2017, 65(3): 554–564.
Related Patents: EP2414528 (B1), US8822146 (B2), US8889352 (B2), US9505797 (B2), EP2414527 (B1), LT5706 (B), US9988673 (B2).
RESEARCH ACTIVITIES in 2020
Precise Genomic Mapping of 5-Hydroxymethylcytosine via Covalent Tether-Directed Sequencing
5-hydroxymethylcytosine (5hmC) is the most prevalent of the oxidized forms of 5mC and is implicated in DNA demethylation and transcriptional regulation in different biological settings. Profiling of this relatively scarce genomic modification in clinical samples requires cost-effective high-resolution techniques that avoid harsh chemical treatment. We developed a bisulfite-free approach for 5hmC profiling at single-nucleotide resolution, named hmTOP-seq, which is based on direct sequence readout primed at covalently labelled 5hmC sites from an in situ tethered DNA oligonucleotide. Examination of distinct conjugation chemistries suggested a structural model for the tether-directed nonhomologous polymerase priming enabling theoretical evaluation of suitable tethers at the design stage. The hmTOP-seq procedure was used to construct 5hmC maps in mouse embryonic stem cells, which demonstrated subtle differences in 5hmC distribution at various genomic elements, intron-exon junctions and both strands of protein-coding genes. Collectively, hmTOP-seq provides a new valuable tool for cost-effective and precise identification of 5hmC in characterizing its biological role and epigenetic changes associated with human disease (Gibas et al. PLOS Biol. 2020, 18(4): e3000684).

Correlation between hmTOP-seq library replicates (left) and between hmTOP-seq and bisulfite-based TAB-seq (right)

Bisulfite-Free Approach for Base-Resolution Analysis of Genomic 5-Carboxylcytosine.
Due to an extreme rarity of 5-carboxylcytosine (5caC) in the mammalian genome, investigation of its role poses a considerable challenge. Methods based on bisulfite sequencing have been proposed for genome-wide 5caC analysis. However, bisulfite-based sequencing of scarcely abundant 5caC demands significant experimental and computational resources, increasing sequencing cost. We developed a bisulfite-free approach, caCLEAR, for high-resolution mapping of 5caCGs. The method uses an atypical activity of the methyltransferase eM.SssI to remove a carboxyl group from 5caC, generating unmodified CGs [4], which are localized by uTOP-seq sequencing [5]. Validation of caCLEAR on model DNA systems and mouse embryonic stem cells supports the suitability of caCLEAR for analysis of 5caCGs. The constructed 5caCG profiles of naive and primed for development pluripotent mouse embryonic stem cells reflect their distinct demethylation dynamics and demonstrate an association of 5caC with gene expression and cell state. We demonstrate that caCLEAR is a robust economical approach that could help provide deeper insights into biological roles of 5caC (Ličytė et al. Cell Rep. 2020, 32: 108155).

caCLEAR approach.
Identification of Foetal Unmodified and 5-Hydroxymethylated CG Sites in Maternal Cell-free DNA for Non-Invasive Prenatal Testing
Numerous studies have demonstrated the presence of cell-free DNA (cfDNA) in various body fluids, which can be used for non-invasive detection of cancer and other diseases or states. Massively parallel sequencing of maternal cfDNA is widely used to test foetal genetic abnormalities in non-invasive prenatal testing. Building upon previous knowledge that placenta, the main source of foetal circulating DNA, is hypomethylated in comparison to maternal tissue counterparts of cfDNA, we propose that targeting either unmodified or 5-hydroxymethylated CG sites specifically enriches foetal genetic material and reduces the numbers of required analytical sequencing reads, thereby decreasing the cost of a test. Using uTOP-seq [5] and hmTOP-seq [Gibas et al. 2020] approaches we constructed uCG and 5hmCG maps of foetal chorionic villi tissue and maternal cfDNA samples which demonstrated that, in contrast to conventional whole genome sequencing, such epigenomic analysis highly specifically enriches foetal DNA fragments from maternal cfDNA. While both our approaches yielded 100% accuracy in detecting Down syndrome in foetuses, hmTOP-seq maintained such accuracy at ultra-low sequencing depths using only one million reads. Robust covalent derivatization followed by targeted analysis of foetal cfDNA by sequencing or qPCR presents an attractive strategy that could help achieve superior sensitivity and specificity in prenatal diagnostics (Gordevičius et al. Clin. Epigenetics. 2020, 12: 153).

Targeted and genome-wide analysis of epigenomic CG sites in liquid biopsies.


