|
|
VYTAUTĖ STARKUVIENĖ-ERFLE
|
|
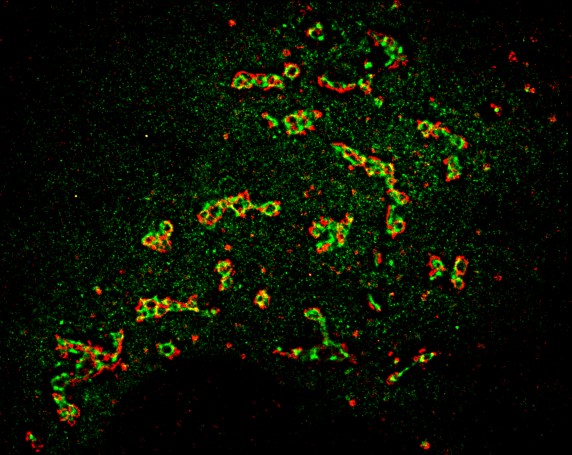
Intracellular trafficking distributes newly synthesized and endocytosed material to diverse cellular destinations and, by doing so, ensures cellular homeostasis. The functionality of the secretory trafficking and endocytosis are regulated in a highly complex manner with hundreds of molecular machineries and multiple pathways acting simultaneously. A precise coordination of transport carriers’ formation, directionality of their movement, fusion to the acceptor membranes and the morphology of intracellular organelles is tightly regulated in a temporal and spatial manner. Trafficking is also closely linked to multitude of other cellular processes: autophagy, cell death, regulation of transcription and translation. Deregulation of cargo trafficking leads to ever-increasing list of such diseases as cancer, cardio-vascular or neurodegenerative ones. Regulation of trafficking as a cellular response function to changes in the surrounding is little understood, and I am especially interested to understand this relation in detail.
To dissect the complexity of trafficking and signalling pathways, we use fluorescent microscopy-based assays, cell biology and biochemistry techniques to identify novel intracellular and extracellular regulators, inter-connections among them. We modify gene, transcript and protein expression function by geneediting, RNA interference and antibody-mediated approaches, respectively. We develop techniques to perform these experiments in highresolution and on a largescale, thereby, extracting high-content information from varying biological scales.
SELECTED PUBLICATIONS
- Grigaitis, P., Starkuviene, V., Rost, U., Serva, A., Pucholt, P., Kummer, U. miRNA target identification and prediction as a function of time in gene expression data. RNA Biology. 2020, Apr 22: 1–11.
- Liu., S. J., Majeed, W., Grigaitis, P., Betts, M. J., Climer, L. K., Starkuviene, V., Storrie., B. Epistatic analysis of the contribution of rabs and kifs to CATCHR family dependent Golgi organization. * Front Cell Dev Bio. 2019, Aug 2,7: 126.
- Starkuviene, V., Kallenberger, S. M., Beil, N., Lisauskas, T., Schumacher, B. S., Bulkescher, R., Wajda, P., Gunkel, M., Beneke, J., Erfle, H. High-density cell arrays for genome-scale phenotypic screening. SLAS Discov. 2019 Mar, 24(3): 274–283.
- Bulkescher, R., Starkuviene, V., & Erfle, H. Solid-phase reverse transfection for intracellular delivery of functionally active proteins. Genome Res. 2017 Oct, 27(10): 1752–1758.
- Gunkel, M., Erfle, H., & Starkuviene, V. High-content analysis of the Golgi complex by correlative screening microscopy. Methods Mol Biol. 2016, 111–121.