Biological Modification of DNA and RNA
Epigenetic Modifications of DNA and RNA in Mammals
In recent years, epigenetic phenomena have become a major focus in studies of embryonic development, genomic imprinting and complex human diseases. One of the best-understood epigenetic mechanisms is enzymatic DNA methylation. In the mammalian genome, cytosines in CpG dinucleotides are often methylated to 5-methylcytosine (m5C), which is brought about by combined action of three known AdoMet–dependent DNA methyltransferases (DNMTs). DNA methylation profiles are highly variable across different genetic loci, cell types and organisms, and are dependent on age, sex, diet and disease. Besides m5C, certain genomic DNAs contain detectable amounts of 5-hydroxymethylcytosine (hmC) and lower levels of 5-formylcytosine and 5-carboxylcytosine (caC), which are produced by the oxidation of m5C residues by TET oxygenases. However, many details of how these modifications are established at specific loci and how they control cellular events remain obscure [1].
More than 160 chemically distinct covalent modifications have been detected across various RNA species in prokaryotic and eukaryotic cells. One of the most abundant and important RNA modifications is methylation of the 2'OH group. miRNAs, piRNAs and siRNAs are small non-coding RNA molecules that control gene activity in a homology-dependent manner. Biogenesis of miRNAs and siRNAs in plants involves a methylation step catalysed by the HEN1 methyltransferase, whereas piRNAs are similarly modified in animals [2,3].
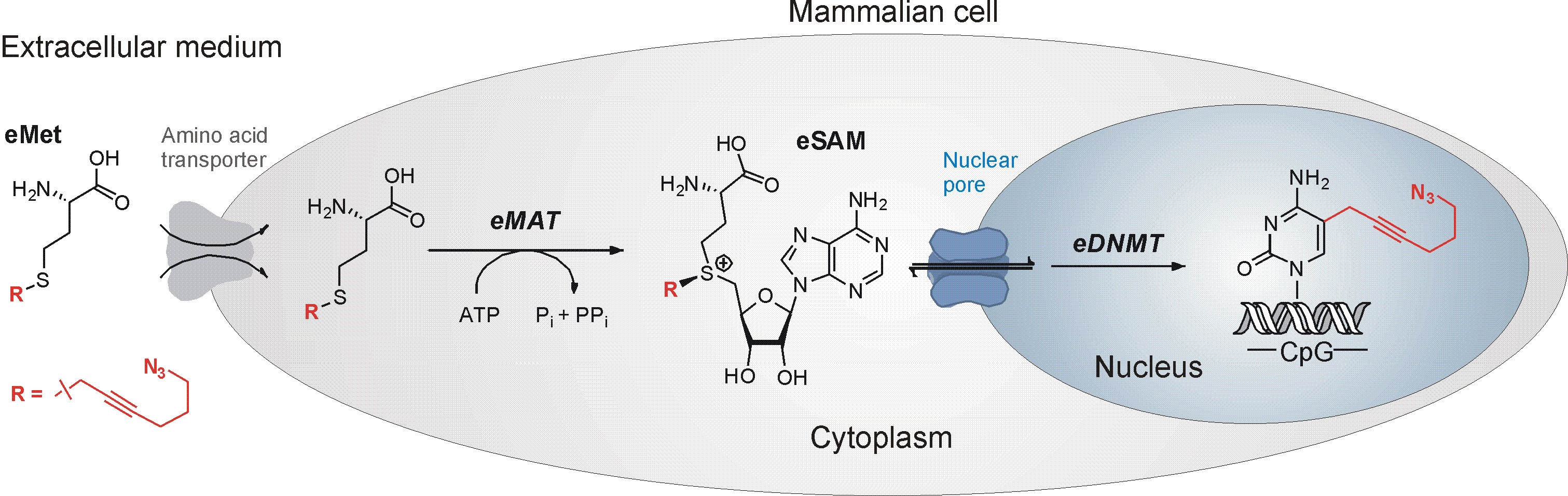
Following our long-standing interest in mechanistic studies of DNA MTases, we turned our focus on advancing DNA and RNA modification analysis and its applications for studies of epigenetic mechanisms [3,4]. Our current ERC-supported studies seek to gain in-depth understanding of how the DNA methylation patterns are established by the three known DNMTs during differentiation and development. Here, our efforts are devoted to devising single-cell methodologies that permit precise determination of where and when the methylation marks are deposited by the individual DNMTs inside living cells (see Figure above).
SELECTED PUBLICATIONS
1. Liutkevičiūtė, Z., Kriukienė, E. Ličytė, J., Rudytė, M., Urbanavičiūtė, G., Klimašauskas, S. Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases. J. Am. Chem. Soc. 2014, 136: 5884–5887.
2. Baranauskė, S., Mickutė, M., Plotnikova, A., Finke, A., Venclovas, Č., Klimašauskas, S., Vilkaitis, G. Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 2015, 43(5): 2802–2812.
3. Mickutė, M., Nainytė, M., Vasiliauskaitė, L., Plotnikova, A., Masevičius, V., Klimašauskas, S., Vilkaitis, G. Animal Hen1 2'-O-methyltransferases as tools for 3'-terminal functionalization and labelling of single-stranded RNAs. Nucleic Acids Res. 2018, 46(17): e104.
4. Kweon, S. M., Chen, Y., Moon, E., Kvederavičiūtė, K., Klimašauskas, S., Feldman, D. E. An adversarial DNA N6-methyladenine-sensor network preserves polycomb silencing. Mol. Cell. 2019, 74: 1138–1147.
5. Tomkuvienė, M., Mickutė, M., Vilkaitis, G., Klimašauskas S. Repurposing enzymatic transferase reactions for targeted labeling and analysis of DNA and RNA. Curr. Opin. Biotechnol. 2019, 55: 114–123.
Enzymatic Hydroxylation and Excision of Extended 5-Methylcytosine Analogues
Methylation of cytosine to 5-methylcytosine (mC) is a prevalent reversible epigenetic mark in vertebrates established by DNA methyltransferases (MTases); the methylation mark can be actively erased via a multi-step demethylation mechanism involving oxidation by Ten-eleven translocation (TET) enzyme family dioxygenases, excision of the latter oxidation products by thymine DNA (TDG) or Nei-like 1 (NEIL1) glycosylases followed by base excision repair to restore the unmodified state. Here we probed the activity of the mouse TET1 (mTET1) and Naegleria gruberi TET (nTET) oxygenases with DNA substrates containing extended derivatives of the 5-methylcytosine carrying linear carbon chains and adjacent unsaturated carbon-carbon bonds. We found that the nTET and mTET1 enzymes were active on modified mC residues in single-stranded and double-stranded DNA in vitro, while the extent of the reactions diminished with the size of the extended group. Iterative rounds of nTET hydroxylations of ssDNA proceeded with high stereo specificity and included not only the natural alpha position but also the adjoining carbon atom in the extended side chain. The regioselectivity of hydroxylation was broken when the reactive carbon was adjacent to an sp1 or sp2 system. We also found that NEIL1 but not TDG was active with bulky TET-oxidation products. These findings provide important insights into the mechanism of these biologically important enzymatic reactions (Tomkuvienė et al. J. Mol. Biol. 2020, 423: 6157–6167).

Photocage-Selective Capture and Light-Controlled Release of Target Proteins
Photochemical transformations enable exquisite spatio-temporal control over biochemical processes, however, methods for reliable manipulations of biomolecules tagged with biocompatible photo-sensitive reporters are lacking. We went on to create a high affinity binder specific to a photolytically removable caging group. We utilized chemical modification or genetically-encoded incorporation of noncanonical amino acids to produce proteins with photocaged cysteine or selenocysteine residues which were used for raising a high affinity monoclonal antibody against a small photoremovable tag, 4,5-dimethoxy-2-nitrobenzyl (DMNB) group. Employing the produced photocage-selective binder, we demonstrate selective detection and immunoprecipitation of a series of DMNB-caged model proteins and DNA cytosine-5 methyltransferase enzymes from complex biological mixtures. The proposed orthogonal strategy permits photocage-selective capture and light-controlled traceless release of target proteins for a myriad of applications in nanoscale assays (Rakauskaitė et al. iScience. 2020, 23(12): 101833; Klimašauskas et al., LT2020539).

Hen1 Methyltransferase-Directed RNA Capture and Sequencing of miRNAs and Bacterial Small RNAs in Probiotic Lactobacillus casei
Targeted installation of designer chemical moieties on biopolymers provides an orthogonal means for their visualization, manipulation and sequence analysis. Although high-throughput RNA sequencing is a widely used method for transcriptome analysis, certain steps, such as 3’ adapter ligation in strand-specific RNA sequencing, remain challenging despite numerous optimizations. Here we remedy this limitation by adapting two small RNA 2’-O-methyltransferases, ssRNA-specific DmHen1 and dsRNA modifying AtHEN1, for orthogonal chemo-enzymatic click tethering of a 3’ sequencing adapter that supports cDNA production by reverse transcription of the tagged RNA. We show by profiling a reference miRNA pool and the small RNA transcriptome of probiotic Lactobacillus casei BL23 that the developed methyltransferase-captured 3’ RNA sequencing technique, meCap-seq, can advance analysis of eukaryotic and prokaryotic ssRNA pools. Our findings provide a valuable resource for studies of the regulatory RNome in Lactobacilli and pave the way to developing novel transcriptome and epitranscriptome profiling approaches in vitro and inside living cells (EP3271478 B1; Mickutė et al., submitted).



