|
|
ROLANDAS MEŠKYS Distinguished Professor Head, Department of Molecular Microbiology and Biotechnology Institute of Biochemistry Email: Phone: +370 5 223 4386 |
|
Microbial Biochemical Diversity as a Source of New Biocatalysts
Modern biotechnology is based on the application of enzymes derived predominantly from microorganisms. Both genetic and biochemical microbial diversity is an immense source of different proteins and biocatalysts. The analysis and exploration of said diversity is one of the main aims of our group. The studies are concentrated on several fields. The first one is related to the isolation of N-heterocyclic compound-utilizing microorganisms and the investigation of the catabolic pathways of these compounds in individual bacteria. Unique oxygenases active towards indole, pyridine and 4-hydroxypyridine as a primary substrate have been characterized, genetically modified and applied for development of biocatalytic processes [1–4]. Screening for novel enzymes is also carried out by applying metagenomic techniques – effective selection systems combined with tailored substrates [5].
More than 160 of differently modified nucleotides play a crucial role in various biological processes. Also, various modified nucleotides are used as promising building blocks for programmable changes of nucleic acids. The biosynthetic pathways of many modified nucleotides including N4-acetylcytosine derivatives are well understood, but the catabolism or salvage of those compounds are only scarcely studied. For the first time, we show that in E. coli the ASCH domain-containing protein YqfB, which has a unique Thr-Lys-Glu catalytic triad, catalyses the hydrolysis of N4-acetylcytidine. In addition, novel N4 amino acid-acylated and alkylated 2'-deoxycytidine analogues were synthesized.
SELECTED PUBLICATIONS
1. Časaitė et al. Microbial degradation of pyridine: a complete pathway deciphered in Arthrobacter sp. 68b. Appl. Environ. Microbiol. 2020, 86: e00902-20.
2. Sadauskas et al. Bioconversion of biologically active indole derivatives with indole-3-acetic acid-degrading enzymes from Caballeronia glathei DSM50014. Biomolecules. 2020, 10: 663.
3. Vaitekūnas et al. Biochemical and genetic analysis of 4-hydroxypyridine catabolism in Arthrobacter sp. strain IN13. Microorganisms. 2020, 8: 888.
4. Sadauskas et al. Enzymatic synthesis of novel water-soluble indigoid compounds. Dyes Pigm. 2020, 173: 107882.
5. Urbelienė et al. A rapid method for the selection of amidohydrolases from metagenomic libraries by applying synthetic nucleosides and a uridine auxotrophic host. Catalysts. 2020, 10: 445.
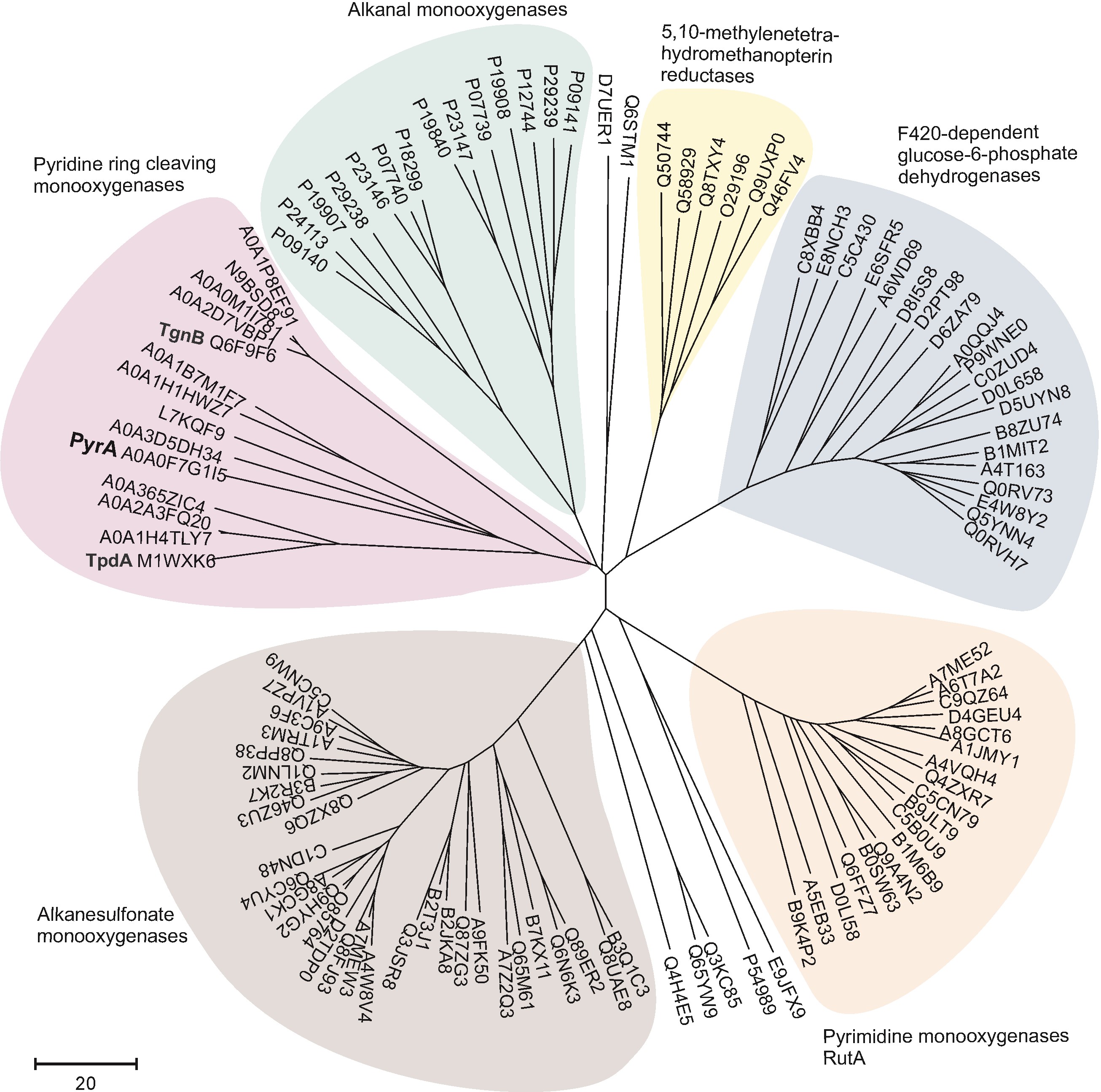
Microbial Degradation of Pyridine: A Complete Pathway Deciphered in Arthrobacter sp. 68b
Pyridine and its derivatives constitute majority of heterocyclic aromatic compounds that occur largely as a result of human activities and contribute to the environmental pollution. It is known that they can be degraded by various bacteria in the environment, however, the degradation of unsubstituted pyridine has not yet been completely resolved. Here, we present data on the pyridine catabolic pathway in Arthrobacter sp. 68b at the level of genes, enzymes and metabolites. The pyr genes cluster, responsible for the degradation of pyridine, was identified in a catabolic plasmid p2MP. The pathway of pyridine metabolism consisted of four enzymatic steps and ended by formation of succinic acid. The first step in the degradation of pyridine proceeds through a direct ring cleavage catalysed by a two-component flavin-dependent monooxygenase system, encoded by pyrA and pyrE genes. The genes pyrB, pyrC, and pyrD were found to encode (Z)-N-(4-oxobut-1-enyl)formamide dehydrogenase, amidohydrolase, and succinate semialdehyde dehydrogenase, respectively. These enzymes participate in the subsequent steps of pyridine degradation (Časaitė et al. Appl. Environ. Microbiol. 2020, 86: e00902-20).

In vitro reconstruction of the pyridine degradation pathway
(a) The analysis of pyridine metabolites by LC-MS and the metabolic pathway of pyridine. Samples were analysed in the positive or negative ionization mode; the extracted ion chromatograms correspond to the quasimolecular ions of (1) pyridine (m/z=80 [M+H]+), metabolite 2 – (Z)-N-(4-oxobut-1-enyl)formamide (m/z=114 [M+H]+), metabolite 3 – (Z)-4-formamidobut-3-enoic acid (m/z=130 [M+H]+), 4 – succinic acid semialdehyde (m/z=101 [M-H]–), and 5 – succinate (m/z=117 [M-H]–);
(b) purified proteins participating in pyridine pathway;
(c) pyr gene cluster in the p2MP plasmid. PyrA – pyridine monooxygenase, PyrE and FR – flavin reductase, PyrB – (Z)-N-(4-oxobut-1-enyl)formamide dehydrogenase, PyrC – amidohydrolase, PyrD – succinate semialdehyde dehydrogenase.
YqfB Protein from Escherichia coli: Atypical Amidohydrolase Active towards N4-acylcytosine Derivatives
Human activating signal cointegrator homology (ASCH) domain-containing proteins are widespread and diverse but, at present, the vast majority of those proteins have no function assigned to them. This study demonstrates that the 103-amino acid hypothetical protein YqfB from E. coli is a unique ASCH domain-containing amidohydrolase responsible for the catabolism of N4-acetylcytidine (ac4C). YqfB has several interesting and unique features: (i) it is the smallest monomeric amidohydrolase, (ii) it is active towards structurally different N4-acylated cytosines/cytidines, iii) it has a very high activity rate (kcat/Km up to 2.8×106 M–1s–1) and (iv) it contains a unique Thr-Lys-Glu catalytic triad and Arg acting as an oxyanion hole. YqfB ability to hydrolyse various N4-acylated cytosines and cytidines not only sheds light on the long-standing mystery of how ac4C is catabolized in bacteria, but also expands our knowledge of the structural diversity within the active sites of amidohydrolases. (Stanislauskienė et al. Sci. Rep. 2020, 10: 788).

Predicted structure of the active centre of YqfB and proposed catalytic mechanism of ac4C hydrolysis
(a) The overall structure of the proposed enzyme-substrate complex.
(b) A detailed view of Yqfb active centre with bound ac4C, as generated by MD simulation; dashed lines correspond to hydrogen bonds.
(c) The acylation step of ac4C hydrolysis catalysed by YqfB.
Bioconversion of Biologically Active Indole Derivatives with Indole-3-Acetic Acid-Degrading Enzymes from Caballeronia glathei DSM50014
A plant auxin hormone indole-3-acetic acid (IAA) can be assimilated by bacteria as an energy and carbon source, although no degradation has been reported for indole-3-propionic acid and indole-3-butyric acid. Caballeronia glathei possesses a full iac gene cluster and is able to use IAA as a sole source of carbon and energy. Next, IacE is responsible for the conversion of 2-oxoindole-3-acetic acid intermediate into the central intermediate 3-hydroxy-2-oxindole-3-acetic acid (DOAA). Finally, IacA and IacE were shown to convert a wide range of indole derivatives, including indole-3-propionic acid and indole-3-butyric acid, into corresponding DOAA homologs. This work provides novel insights into Iac-mediated IAA degradation and demonstrates the versatility and substrate scope of IacA and IacE enzymes. (Sadauskas et al. Biomolecules. 2020, 10: 663).

Comparison of biodegradation of indole in Acinetobacter sp. strain O153 and biodegradation of IAA in Caballeronia glathei DSM50014 (M. Sadauskas, PhD thesis).
Grey reaction arrows indicate spontaneous reactions. Arrows with identical colours indicate genes and proteins with similar predicted functions.