Single-molecule studies of protein and DNA interactions
Real-time monitoring of single-proteins on nucleic acid (NA) substrates is an essential tool for studying NA-interacting proteins allowing better mechanistic understanding. To follow individual proteins on a NA molecule, one or both ends of NA molecule that are extended by an external force are attached to the surface. Over the past 20 years, a number of such methods emerged: tethered particle motion (TPM), optical or magnetic tweezers (OTs or MTs), flow-stretch assays and various combinations of these methods. Stretched NAs is a common means to investigate the dynamics of protein-NA interactions. Several experimental strategies are employed to extend and align NA molecules (e.g. NA combing or through hydrodynamic flow). The hydrodynamic flow method allows attaching NA molecules to the glass surface, where they are stretched by a shear hydrodynamic flow. The flow can be applied to extend single-end tethered NA, or deployed to allow the specific double-end tethering. Anchoring the NA onto a lipid bilayer and a diffusion barrier etched on the microscope slide causes the alignment of the NA moving under a buffer flow. This method, known as DNA curtains, allows imaging of many DNA molecules in parallel.
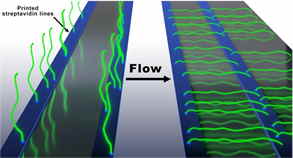
Recently our team developed an alternative assay to the original DNA Curtains that we termed the “Soft” DNA Curtains (LRC grant No. S-MIP-17-59). We fabricated streptavidin patterns (i.e. line-features) on the modified coverslip surface that can be utilized to assemble stably immobilized biotinylated DNA arrays. The application of hydrodynamic buffer flow allows extension of the immobilized DNA molecules along the surface of the flow cell channel. We also fabricated the uniformly oriented double-tethered DNA Curtains using heterologous labelling of the DNA by biotin and digoxigenin. We increased the stability of the immobilized DNA molecules using a more stable alternative to sAv called traptavidin as an ink for the fabrication of protein templates.
The main goal of our research topic is to apply the developed platform for studies of DNA targeting mechanisms of diverse CRISPR-Cas systems family (LRC grant No. S-MIP-20-55), novel molecular-tools – prokaryotic Argonaute (pAgo) proteins (LRC grant No. 09.3.3-LMT-K-712-19-0113) and various restriction endonucleases. Our newest publication on the “Soft” DNA Curtains [1] and its continuation [2] received a broad interest of scientists from various fields.
SELECTED PUBLICATIONS
- Tutkus, M., Rakickas, T., Kopūstas, A., Ivanovaitė, Š., Venckus, O., Navikas, V., Zaremba, M., Manakova, E., Valiokas, R. Fixed DNA molecule arrays for high-throughput single DNA-protein interaction studies. Langmuir. 2019, 35, 17: 5921–5930.
- Kopūstas, A., Ivanovaitė, Š., Rakickas, T., Pocevičiūtė, E., Paksaitė, J., Karvelis, T., Zaremba, M., Manakova, E., Tutkus, M. bioRxiv. doi.org/10.1101/2020.06.15.151662 (in press).
- Tutkus, M., Marciulionis, T., Sasnauskas, G., Rutkauskas, D. DNA-endonuclease complex dynamics by simultaneous FRET and fluorophore intensity in evanescent field. Biophysical Journal. 2017, 112(5): 850–858.
- Tutkus, M., Sasnauskas, G., Rutkauskas, D. Probing the dynamics of restriction endonuclease NgoMIV-DNA interaction by single-molecule FRET. Biopolymers. 2017, 107(12): e23075.


